---
title: "Parameters and Optimization in TVB-Optim"
format:
html:
code-fold: false
toc: true
toc-depth: 3
fig-width: 8
out-width: "100%"
jupyter: python3
execute:
cache: true
---
# Why Parameters?
Parameters serve two purposes:
1. Define which values get differentiated and subsequently optimized.
2. Enforce constraints during optimization by applying transformations, eg a positive constraint on a parameter.
## General Properties of Parameters
Parameters define the `__jax_array__` method, which allows us to use them in the same way as jax arrays.
```{python}
import jax
import jax.numpy as jnp
from tvboptim.types import Parameter
p = Parameter(jnp.array([1.0, 2.0, 3.0]))
print("Shape:", p.shape)
print("Numeric op:", p + 1)
print("Jax op:", jnp.sin(p))
```
It is good practice to only use parameters for optimization and collect them afterwards for postprocessing as the `__jax_array__` protocol is still experimental ([More info](https://docs.jax.dev/en/latest/jep/28661-jax-array-protocol.html)) and might lead to unexpected results.
```{python}
from tvboptim.types import collect_parameters
collect_parameters(p) # collect all arrays from the Parameters in a PyTree
```
## Marking Parameters for Optimization
After using `prepare` on an experiment (TVB-O) or network (tvboptim.experimental.network_dynamics), we have a state object which is a PyTree with jax arrays at its leaves. The state helps structuring the many values and inputs needed for the simulation in a simple object for convenience. But in most cases we only want to subject a fraction of the state to optimization. We can mark the values in the state for optimization by wrapping them in the `Parameter` type:
```{python}
from tvboptim.experimental.network_dynamics import Bunch # A dictionary with attribute access
from tvboptim.types import Parameter, show_parameters
# A simple example state
simple_state = Bunch(
global_coupling = 0.1,
time_constant = 1.0,
)
# Mark global coupling for optimization
simple_state.global_coupling = Parameter(simple_state.global_coupling)
show_parameters(simple_state)
```
Inside e.g. `OptaxOptimizer` we can use the knowledge of what to consider a parameter to partition the state into two sub PyTrees, one that gets differentiated and one that stays static:
```{python}
from tvboptim.types import partition_state, combine_state
diff_state, static_state = partition_state(simple_state)
print("Diff state:")
print(diff_state)
print("Static state:")
print(static_state)
```
Sub PyTrees can be combined back to a single PyTree using `combine_state`. This is something that the `OptaxOptimizer` does automatically.
```{python}
#| eval: false
# Schematics of the `OptaxOptimizer`
def loss_wrapper(diff_state, static_state):
state = combine_state(diff_state, static_state) # Recombine parameters with static values
return loss(state)
jax.grad(loss_wrapper)(diff_state, static_state) # grad by default acts only on the first argument
```
## Enforcing Constraints
Some values in a model can be constrained through their physiological meaning. For example, we want the global coupling to be positive. The optimizer itself does not know about these constraints, so we have to apply them explicitly.
```{python}
from tvboptim.types import BoundedParameter
p1 = Parameter(-0.0001)
p2 = BoundedParameter(0.5, low=0.0, high=1.0)
p3 = BoundedParameter(-0.0001, low=0.0, high=1.0)
# Simple loss that would increase p to infinity in minimization
def loss(p):
return -p
print("Gradient without constraint:", jax.grad(loss)(p1))
print("Gradient with fulfilled constraint:", jax.grad(loss)(p2))
print("Gradient with violated constraint:", jax.grad(loss)(p3))
```
We provide several predefined parameter types:
* `Parameter`
* `BoundedParameter`
* `NormalizedParameter`
* `TransformedParameter`
* `SigmoidBoundedParameter`
* `MaskedParameter`
Custom constraints can be implemented as well, for that either `Parameter` can be subclassed directly or we use `TransformedParameter` and define our constraint as forward and reverse transforms. As an example lets look at how we would build a sigmoid bounded parameter. This has the advantage over `BoundedParameter` that we always get a gradient, while the `BoundedParameter` returns 0 when the constraint is violated and it then stays at the lower or upper bound indefinitely.
```{python}
from tvboptim.types import TransformedParameter, SigmoidBoundedParameter
def sigmoid_bounded_param(value, low=0.0, high=1.0):
def forward(x):
return low + (high - low) * jax.nn.sigmoid(x)
def inverse(x):
normalized = (x - low) / (high - low)
return jnp.log(normalized / (1 - normalized))
return TransformedParameter(value, forward, inverse)
# Check if the constraint is fulfilled
p_custom = sigmoid_bounded_param(0.5, low=0.0, high=1.0)
print("Custom TransformedParameter:", p_custom)
print("Gradient with smooth constraint:", jax.grad(loss)(p_custom))
```
To check if the constraint is fulfilled we run an Optimization on the simple loss function, the parameter should stay below 1:
```{python}
from tvboptim.optim import OptaxOptimizer
import optax
opt = OptaxOptimizer(loss, optax.adam(0.01))
p_opt, _ = opt.run(p_custom, max_steps=1000)
print("Optimized parameter:", p_opt)
print("Smaller than 1:", p_opt < 1.0)
```
# A Simple Example
As a simple example on how to use parameters we try to find the fixed point of the reduced Wong-Wang model. This example demonstrates the key concepts of parameter optimization in TVB-Optim by finding parameter values where the neural activity reaches equilibrium.
## Setting up the Model
First, we load the reduced Wong-Wang model and examine its structure:
```{python}
from tvboptim.experimental.network_dynamics.dynamics import ReducedWongWang
m = ReducedWongWang()
dfun = m.dynamics
m.DEFAULT_PARAMS
```
The model includes parameters like `gamma` (kinetic parameter) and `tau_s` (time constant) that control the dynamics.
## Defining the Optimization Problem
For a specific initial condition and coupling, we have a fixed point if the derivative is zero. We define our loss function to minimize the absolute sum of derivatives:
```{python}
p_init = m.DEFAULT_PARAMS
def loss_fixed_point(p, S_0 = [0.8], coupling = Bunch(instant = [0.0], delayed = [0.0])):
return jnp.abs(jnp.sum(dfun(0.0, S_0, p, coupling, coupling)[0]))
print(f"Initial loss: {loss_fixed_point(p_init):.6f}")
```
Without parameter constraints, JAX would differentiate all parameters in the model:
```{python}
gradients = jax.grad(loss_fixed_point)(p_init)
print("Gradients for all parameters:", gradients)
```
## Running the Optimization
Now we'll set up and run the optimization using both regular and normalized parameters to compare their performance.
### Setting up the Optimizer
We configure the optimizer with callbacks to track progress and automatically stop when convergence is achieved:
```{python}
from tvboptim.optim import OptaxOptimizer, DefaultPrintCallback, StopLossCallback, SavingLossCallback, SavingParametersCallback, MultiCallback
import optax
# Configure callbacks for monitoring optimization progress
cbs = MultiCallback([
DefaultPrintCallback(every=10), # Print progress every 10 steps
SavingLossCallback(), # Save loss history
SavingParametersCallback(), # Save parameter history
StopLossCallback(stop_loss=1e-6), # Stop when loss is sufficiently small
])
opt = OptaxOptimizer(loss_fixed_point, optax.adam(0.01), callback=cbs)
```
### Optimization with Regular Parameters
First, let's optimize using regular `Parameter` objects, focusing only on `gamma` and `tau_s`:
```{python}
# Mark specific parameters for optimization
p_init.gamma = Parameter(p_init.gamma)
p_init.tau_s = Parameter(p_init.tau_s)
print("Starting optimization with regular Parameters...")
p_opt, fitting_data = opt.run(p_init, max_steps=1000)
print(f"Final loss: {fitting_data['loss'].save.values[-1]:.8f}")
```
### Optimization with Normalized Parameters
Now let's compare with `NormalizedParameter`, which can lead to better convergence by normalizing parameter scales (gamma = 0.681 -> 1.0, tau_s = 100 -> 1.0):
```{python}
from tvboptim.types import NormalizedParameter
# Reset and use normalized parameters
p_init.gamma = NormalizedParameter(p_init.gamma)
p_init.tau_s = NormalizedParameter(p_init.tau_s)
print("Starting optimization with NormalizedParameters...")
p_opt_norm, fitting_data_norm = opt.run(p_init, max_steps=1000)
print(f"Final loss: {fitting_data_norm['loss'].save.values[-1]:.8f}")
```
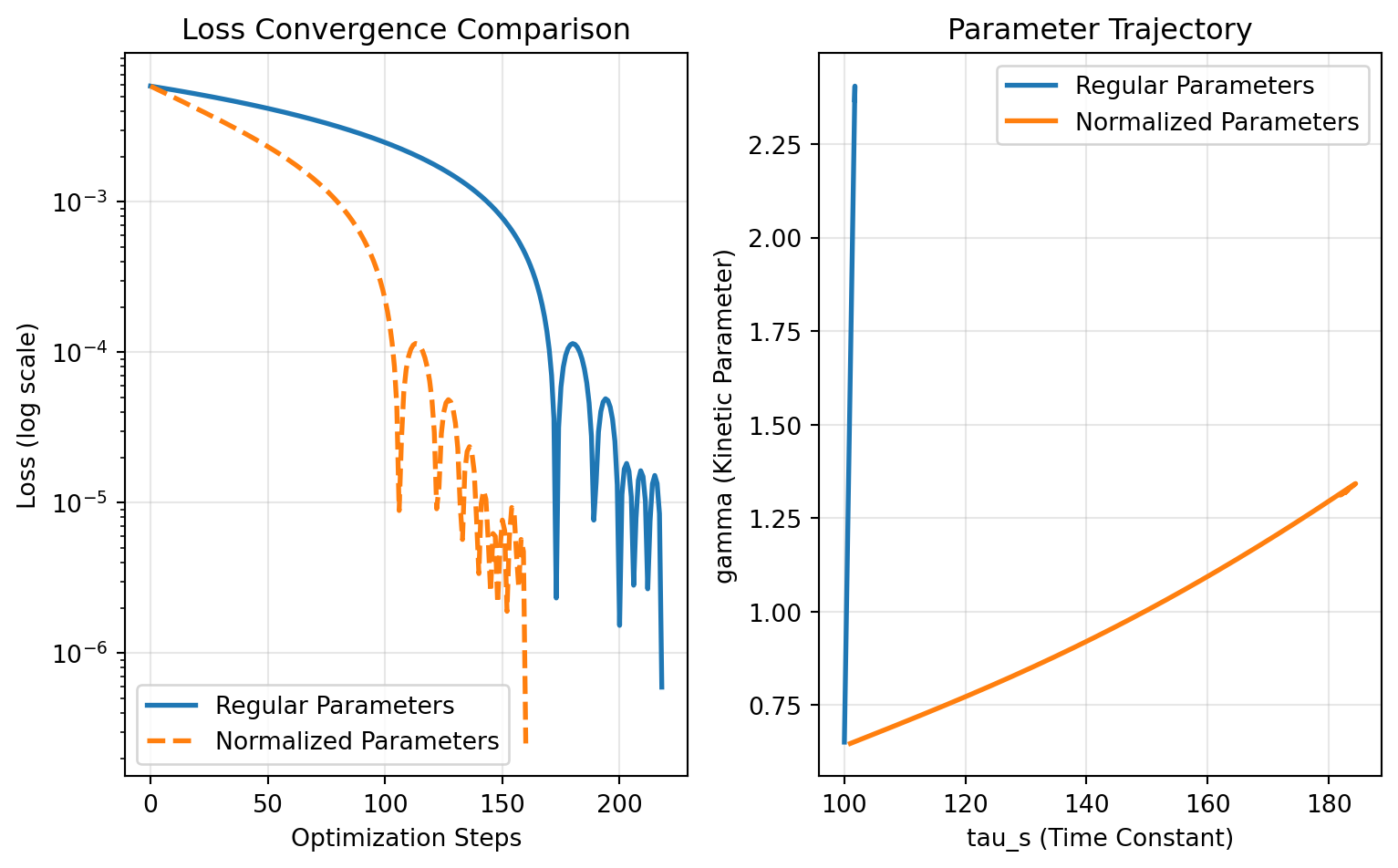
## Comparing Optimization Performance
The normalized parameters converge faster than regular parameters. With regular `Parameter` objects alone, most of the optimization change was applied to `gamma` while `tau_s` barely changed due to scaling differences and a learning rate of 0.01.
### Visualization of Convergence
Let's create plots to visualize the optimization convergence and parameter trajectories:
```{python}
#| code-fold: true
#| code-summary: "Show visualization code"
import matplotlib.pyplot as plt
fig, (ax1, ax2) = plt.subplots(1, 2, figsize=(8.1, 5.0625))
# Loss convergence comparison
ax1.semilogy(fitting_data["loss"].save, label='Regular Parameters', linewidth=2)
ax1.semilogy(fitting_data_norm["loss"].save, label='Normalized Parameters', linewidth=2, linestyle='--')
ax1.set_xlabel('Optimization Steps')
ax1.set_ylabel('Loss (log scale)')
ax1.set_title('Loss Convergence Comparison')
ax1.legend()
ax1.grid(True, alpha=0.3)
# Parameter trajectory in 2D space
gamma_traj = collect_parameters([p.gamma for p in fitting_data["parameters"].save])
tau_s_traj = collect_parameters([p.tau_s for p in fitting_data["parameters"].save])
gamma_traj_norm = collect_parameters([p.gamma for p in fitting_data_norm["parameters"].save])
tau_s_traj_norm = collect_parameters([p.tau_s for p in fitting_data_norm["parameters"].save])
ax2.plot(tau_s_traj, gamma_traj, label='Regular Parameters', linewidth=2)
ax2.plot(tau_s_traj_norm, gamma_traj_norm, label='Normalized Parameters', linewidth=2)
ax2.set_xlabel('tau_s (Time Constant)')
ax2.set_ylabel('gamma (Kinetic Parameter)')
ax2.set_title('Parameter Trajectory')
ax2.legend()
ax2.grid(True, alpha=0.3)
plt.tight_layout()
plt.show()
```
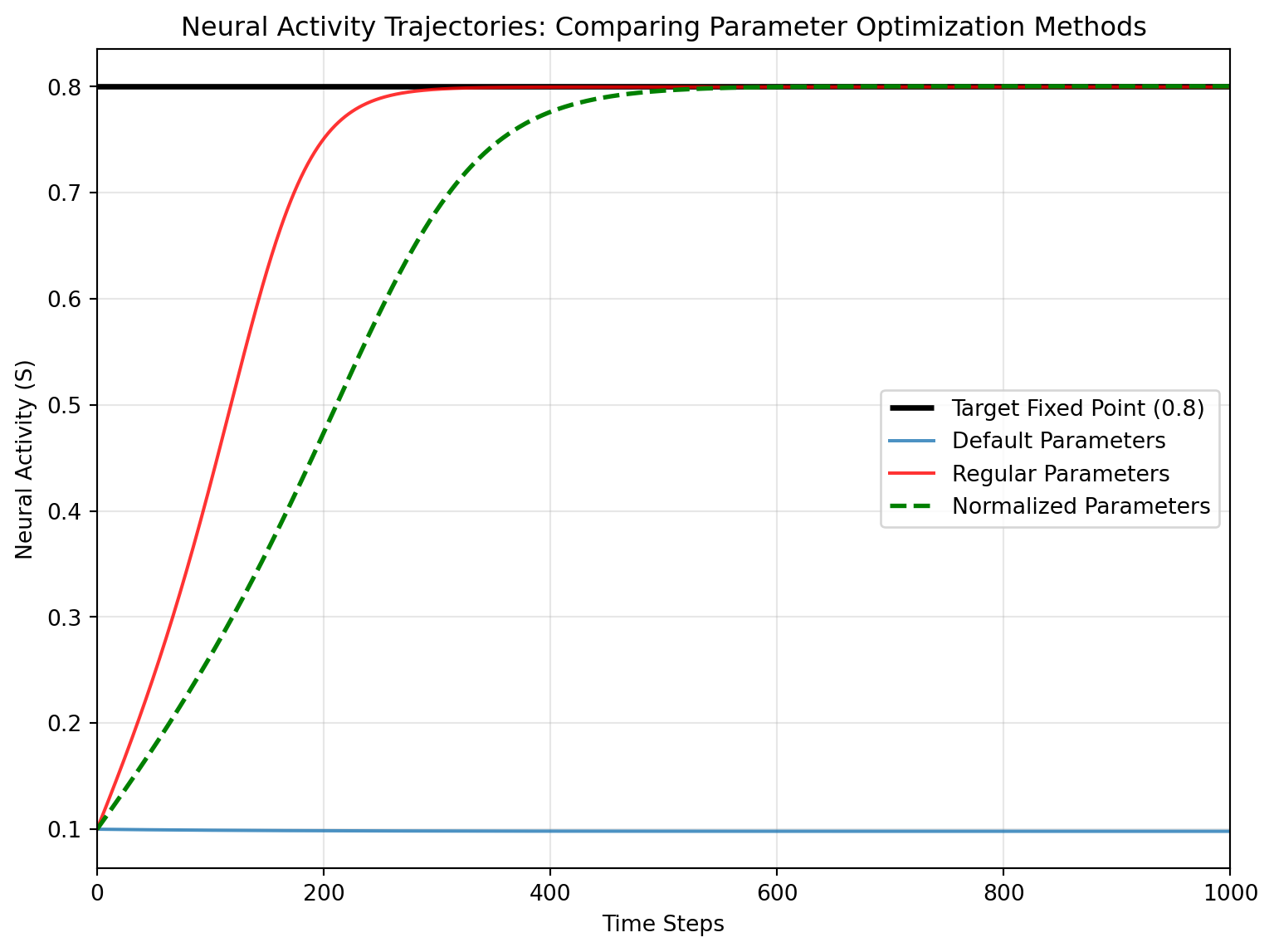
### Validating the Fixed Point Solution
Finally, let's run simulations with the optimized parameters to verify that we've achieved the desired fixed point at 0.8:
```{python}
# Run simulation with default parameters
m = ReducedWongWang()
ts_default = m.simulate(t1 = 1000, dt = 1.0)
# Run simulation with optimized parameters (regular Parameter)
p_opt = collect_parameters(p_opt)
m = ReducedWongWang(gamma = p_opt.gamma, tau_s = p_opt.tau_s)
ts_opt = m.simulate(t1 = 1000, dt = 1.0)
# Run simulation with optimized normalized parameters
p_opt_norm = collect_parameters(p_opt_norm)
m = ReducedWongWang(gamma = p_opt_norm.gamma, tau_s = p_opt_norm.tau_s)
ts_opt_norm = m.simulate(t1 = 1000, dt = 1.0)
```
Now let's visualize how well each optimization approach achieved the target fixed point:
```{python}
#| code-fold: true
#| code-summary: "Show visualization code"
fig, ax = plt.subplots(figsize=(8.1, 6.075))
# Plot target line
ax.hlines(0.8, 0, 1000, color="black", linewidth=2.5, label="Target Fixed Point (0.8)")
# Plot time series
ax.plot(ts_default[1][:, 0, :], label="Default Parameters", alpha=0.8)
ax.plot(ts_opt[1][:, 0, :], color='red', label="Regular Parameters", alpha=0.8)
ax.plot(ts_opt_norm[1][:, 0, :], color='green', linestyle='--',
label="Normalized Parameters", linewidth=2)
ax.set_xlabel('Time Steps')
ax.set_ylabel('Neural Activity (S)')
ax.set_title('Neural Activity Trajectories: Comparing Parameter Optimization Methods')
ax.legend(loc="right")
ax.grid(True, alpha=0.3)
ax.set_xlim(0, 1000)
plt.tight_layout()
plt.show()
```
This visualization shows how the different parameter optimization approaches perform in achieving the desired fixed point.