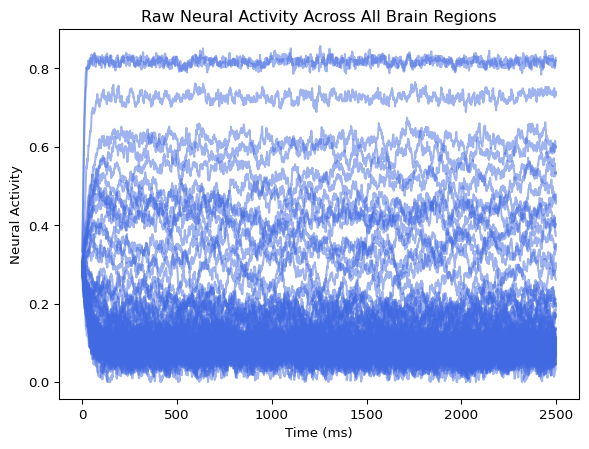
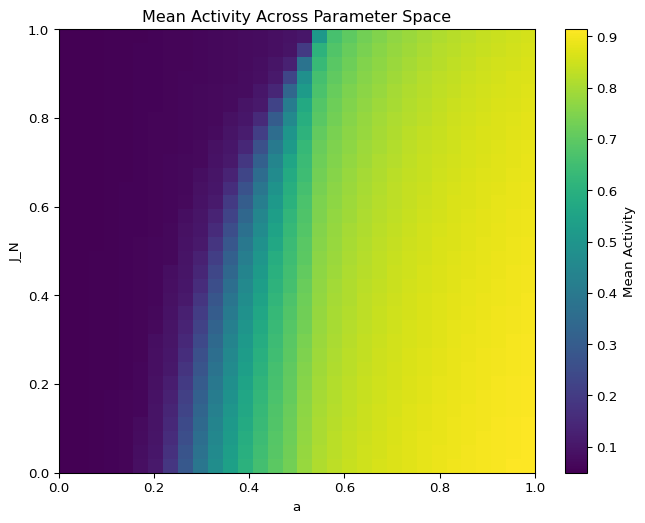
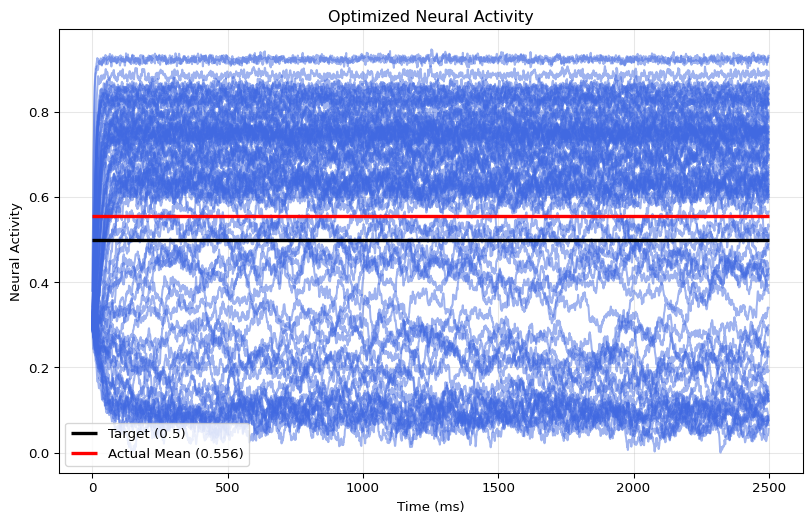
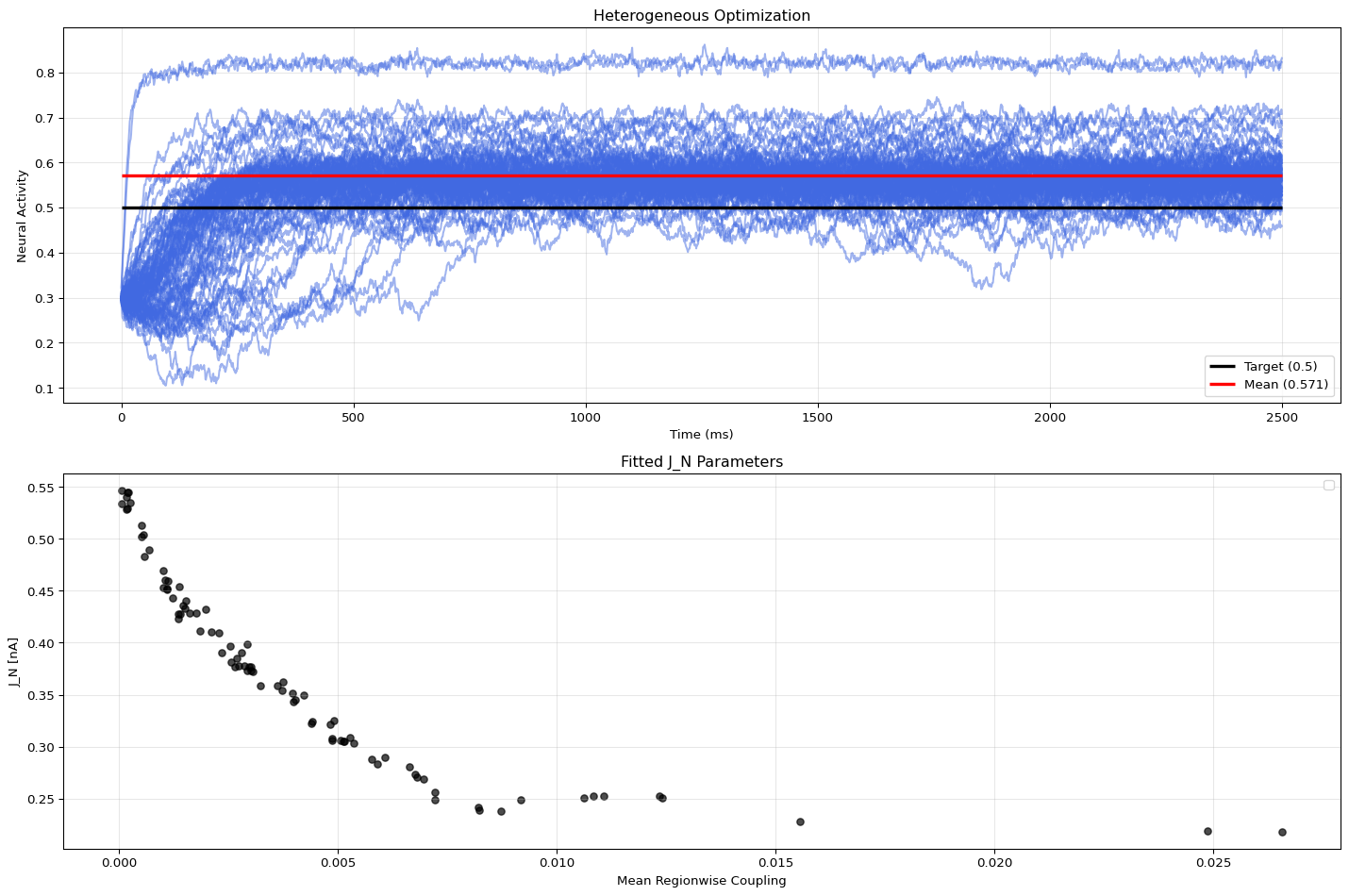
001
002 import jax.scipy.signal as sig
003 from collections import namedtuple
004 import jax
005 from tvbo.data.types import TimeSeries
006 import jax.numpy as jnp
007 import jax.scipy as jsp
008
009
010 def cfun(weights, history, current_state, p, delay_indices, t):
011 n_node = weights.shape[0]
012 a, b = p.a, p.b
013
014 x_j = jnp.array([
015
016 current_state[0],
017
018 ])
019
020 pre = x_j
021
022 def op(x): return weights @ x
023 gx = jax.vmap(op, in_axes=0)(pre)
024 return b + a*gx
025
026
027 def dfun(current_state, cX, _p, local_coupling=0):
028 w, I_o, J_N, a, b, d, gamma, tau_s = _p.w, _p.I_o, _p.J_N, _p.a, _p.b, _p.d, _p.gamma, _p.tau_s
029 # unpack coupling terms and states as in dfun
030 c_pop0 = cX[0]
031
032 S = current_state[0]
033
034 # compute internal states for dfun
035 x = I_o + J_N*c_pop0 + J_N*S*local_coupling + J_N*S*w
036 H = (-b + a*x)/(1 - jnp.exp(-d*(-b + a*x)))
037
038 return jnp.array([
039 -S/tau_s + H*gamma*(1 - S), # S
040 ])
041
042
043 def integrate(state, weights, dt, params_integrate, delay_indices, external_input):
044 """
045 Heun Integration
046 ================
047 """
048 t, noise = external_input
049
050 params_dfun, params_cfun, params_stimulus = params_integrate
051
052 history, current_state = state
053 stimulus = 0
054
055 inf = jnp.inf
056 min_bounds = jnp.array([[[0.0]]])
057 max_bounds = jnp.array([[[1.0]]])
058
059 cX = jax.vmap(cfun, in_axes=(None, -1, -1, None, None, None), out_axes=-
060 1)(weights, history, current_state, params_cfun, delay_indices, t)
061
062 dX0 = dfun(current_state, cX, params_dfun)
063
064 X = current_state
065
066 # Calculate intermediate step X1
067 X1 = X + dX0 * dt + noise + stimulus * dt
068 X1 = jnp.clip(X1, min_bounds, max_bounds)
069
070 # Calculate derivative X1
071 dX1 = dfun(X1, cX, params_dfun)
072 # Calculate the state change dX
073 dX = (dX0 + dX1) * (dt / 2)
074 next_state = current_state + (dX) + noise
075 next_state = jnp.clip(next_state, min_bounds, max_bounds)
076
077 return (history, next_state), next_state
078
079
080 timeseries = namedtuple("timeseries", ["time", "trace"])
081
082
083 def monitor_raw_0(time_steps, trace, params, t_offset=0):
084 dt = 4.0
085 return TimeSeries(time=(time_steps + t_offset) * dt, data=trace, title="Raw")
086
087
088 def monitor_temporal_average_1(time_steps, trace, params, t_offset=0):
089 dt = 4.0
090 voi = jnp.array([0])
091 istep = 1
092 t_map = time_steps[::istep] - 1
093
094 def op(ts):
095 start_indices = (ts,) + (0,) * (trace.ndim - 1)
096 slice_sizes = (istep,) + voi.shape + trace.shape[2:]
097 return jnp.mean(jax.lax.dynamic_slice(trace[:, voi, :], start_indices, slice_sizes), axis=0)
098 vmap_op = jax.vmap(op)
099 trace_out = vmap_op(t_map)
100
101 idxs = jnp.arange(((istep - 2) // 2), time_steps.shape[0], istep)
102 return TimeSeries(time=(time_steps[idxs]) * dt, data=trace_out[0:idxs.shape[0], :, :], title="TemporalAverage")
103
104
105 exp, sin, sqrt = jnp.exp, jnp.sin, jnp.sqrt
106
107
108 def monitor_bold_1(time_steps, trace, params, t_offset=0):
109 # downsampling via temporal average / subsample
110 dt = 4.0
111 voi = jnp.array([0])
112 period = 1000.0 # sampling period of the BOLD Monitor in ms
113 istep_int = 1 # steps taken by the averaging/subsampling monitor to get an interim period of 4 ms
114 istep = 250
115 final_istep = 250 # steps to take on the downsampled signal
116
117 res = monitor_temporal_average_1(time_steps, trace, None)
118 time_steps_i = res.time
119 trace_new = res.data
120
121 time_steps_new = time_steps[jnp.arange(
122 istep-1, time_steps.shape[0], istep)]
123
124 # hemodynamic response function
125 tau_s = params.tau_s
126 tau_f = params.tau_f
127 k_1 = params.k_1
128 V_0 = params.V_0
129 stock = params.stock
130
131 trace_new = jnp.vstack([stock, trace_new])
132
133 def op(var): return 1/3. * exp(-0.5*(var / tau_s)) * (sin(sqrt(1. /
134 tau_f - 1./(4.*tau_s**2)) * var)) / (sqrt(1./tau_f - 1./(4.*tau_s**2)))
135 stock_steps = 5000
136 stock_time_max = 20.0 # stock time has to be in seconds
137 stock_time_step = stock_time_max / stock_steps
138 stock_time = jnp.arange(0.0, stock_time_max, stock_time_step)
139 hrf = op(stock_time)
140
141 # Convolution along time axis
142 # via fft
143 def op1(x): return sig.fftconvolve(x, hrf, mode="valid")
144
145 def op2(x): return jax.vmap(op1, in_axes=(
146 1), out_axes=(1))(x) # map over nodes
147 def op3(x): return jax.vmap(op2, in_axes=(1), out_axes=(1))(
148 x) # map over state variables
149 bold = jax.vmap(op3, in_axes=(3), out_axes=(3))(
150 trace_new) # map over modes
151
152 bold = k_1 * V_0 * (bold - 1.0)
153
154 bold_idx = jnp.arange(
155 final_istep-2, time_steps_i.shape[0], final_istep)[0:time_steps_new.shape[0]] + 1
156 return TimeSeries(time=(time_steps_new + t_offset) * dt, data=bold[bold_idx, :, :], title="BOLD")
157
158
159 def transform_parameters(_p):
160 w, I_o, J_N, a, b, d, gamma, tau_s = _p.w, _p.I_o, _p.J_N, _p.a, _p.b, _p.d, _p.gamma, _p.tau_s
161
162 return _p
163
164
165 c_vars = jnp.array([0])
166
167
168 def kernel(state):
169 # problem dimensions
170 n_nodes = 87
171 n_svar = 1
172 n_cvar = 1
173 n_modes = 1
174 nh = 1
175
176 # history = current_state
177 current_state, history = (state.initial_conditions.data[-1], None)
178
179 ics = (history, current_state)
180 weights = state.connectivity.weights
181
182 dn = jnp.arange(n_nodes) * jnp.ones((n_nodes, n_nodes)).astype(int)
183 idelays = jnp.round(state.connectivity.lengths /
184 state.connectivity.metadata.conduction_speed.value / state.dt).astype(int)
185 di = -1 * idelays - 1
186 delay_indices = (di, dn)
187
188 dt = state.dt
189 nt = state.nt
190 time_steps = jnp.arange(0, nt)
191
192 key = jax.random.PRNGKey(state.noise.metadata.seed)
193 _noise = jax.random.normal(key, (nt, n_svar, n_nodes, n_modes))
194 noise = (jnp.sqrt(dt) * state.noise.sigma[None, ..., None, None]) * _noise
195
196 p = transform_parameters(state.parameters.model)
197 params_integrate = (p, state.parameters.coupling, state.stimulus)
198
199 def op(ics, external_input): return integrate(ics, weights,
200 dt, params_integrate, delay_indices, external_input)
201
202 latest_carry, res = jax.lax.scan(op, ics, (time_steps, noise))
203
204 trace = res
205
206 t_offset = 0
207 time_steps = time_steps + 1
208
209 params_monitors = state.monitor_parameters
210 result = [monitor_raw_0(time_steps, trace, params_monitors[0], t_offset=t_offset),
211 monitor_bold_1(time_steps, trace,
212 params_monitors[1], t_offset=t_offset),
213 ]
214
215 return result