Akil, Huda, Maryann E. Martone, and David C. Van Essen. 2011.
“Challenges and Opportunities in Mining Neuroscience Data.” Science 331 (6018): 708–12.
https://doi.org/10.1126/science.1199305.
Botvinik-Nezer, Rotem, Felix Holzmeister, Colin F. Camerer, Anna Dreber, Juergen Huber, Magnus Johannesson, Michael Kirchler, et al. 2020.
“Variability in the Analysis of a Single Neuroimaging Dataset by Many Teams.” Nature 582 (7810): 84–88.
https://doi.org/10.1038/s41586-020-2314-9.
Cabrera-Álvarez, Jesús, Leon Stefanovski, Leon Martin, Gianluca Susi, Fernando Maestú, and Petra Ritter. 2024.
“A Multiscale Closed-Loop Neurotoxicity Model of Alzheimer’s Disease Progression Explains Functional Connectivity Alterations.” Eneuro 11 (4): ENEURO.0345–23.2023.
https://doi.org/10.1523/eneuro.0345-23.2023.
Deco, Gustavo, Josephine Cruzat, Joana Cabral, Gitte M. Knudsen, Robin L. Carhart-Harris, Peter C. Whybrow, Nikos K. Logothetis, and Morten L. Kringelbach. 2018.
“Whole-Brain Multimodal Neuroimaging Model Using Serotonin Receptor Maps Explains Non-Linear Functional Effects of LSD.” Current Biology 28 (19): 3065–3074.e6.
https://doi.org/10.1016/j.cub.2018.07.083.
Desikan, Rahul S., Florent Ségonne, Bruce Fischl, Brian T. Quinn, Bradford C. Dickerson, Deborah Blacker, Randy L. Buckner, et al. 2006.
“An Automated Labeling System for Subdividing the Human Cerebral Cortex on MRI Scans into Gyral Based Regions of Interest.” NeuroImage 31 (3): 968–80.
https://doi.org/10.1016/j.neuroimage.2006.01.021.
Fischl, Bruce, David H. Salat, Evelina Busa, Marilyn Albert, Megan Dieterich, Christian Haselgrove, Andre van der Kouwe, et al. 2002.
“Whole Brain Segmentation.” Neuron 33 (3): 341–55.
https://doi.org/10.1016/s0896-6273(02)00569-x.
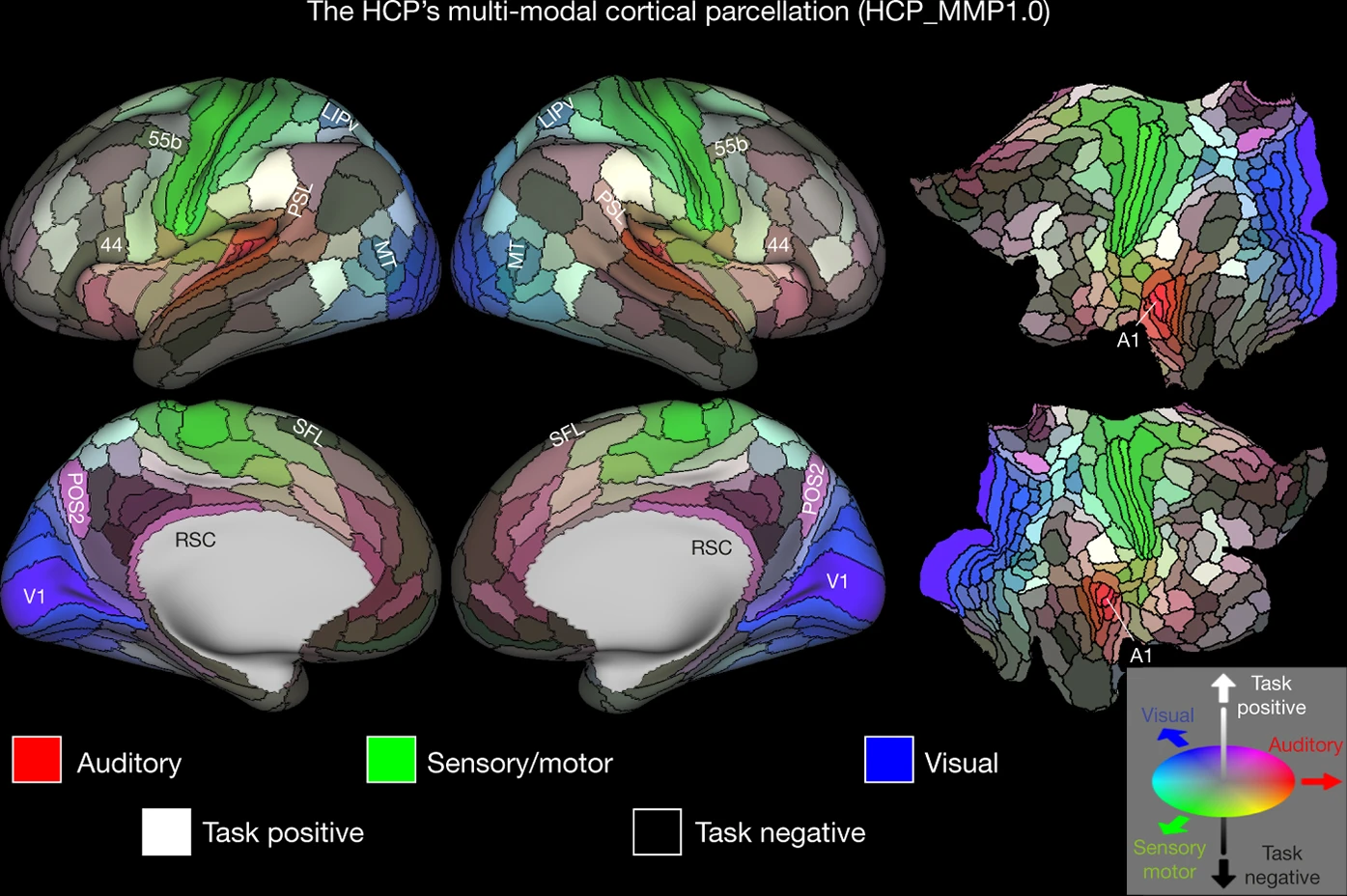
Glasser, Matthew F., Timothy S. Coalson, Emma C. Robinson, Carl D. Hacker, John Harwell, Essa Yacoub, Kamil Ugurbil, et al. 2016.
“A Multi-Modal Parcellation of Human Cerebral Cortex.” Nature 536 (7615): 171–78.
https://doi.org/10.1038/nature18933.
Glasser, Matthew F., Stamatios N. Sotiropoulos, J. Anthony Wilson, Timothy S. Coalson, Bruce Fischl, Jesper L. Andersson, Junqian Xu, et al. 2013.
“The Minimal Preprocessing Pipelines for the Human Connectome Project.” NeuroImage 80 (October): 105–24.
https://doi.org/10.1016/j.neuroimage.2013.04.127.
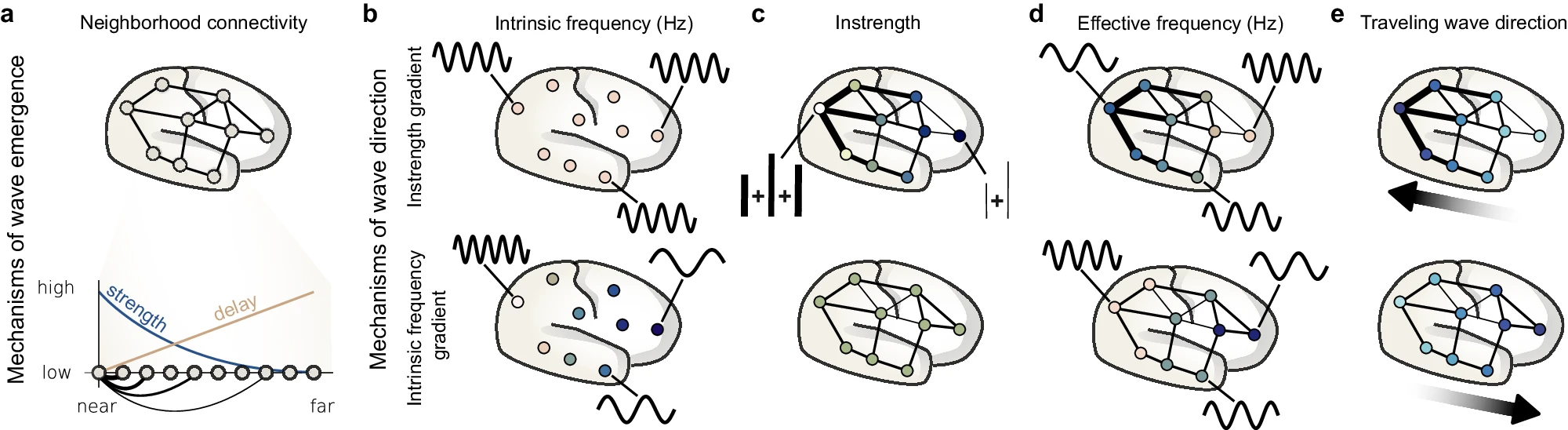
Koller, Dominik P., Michael Schirner, and Petra Ritter. 2024.
“Human Connectome Topology Directs Cortical Traveling Waves and Shapes Frequency Gradients.” Nature Communications 15 (1).
https://doi.org/10.1038/s41467-024-47860-x.
Lindquist, Martin. 2020.
“Neuroimaging Results Altered by Varying Analysis Pipelines.” Nature 582 (7810): 36–37.
https://doi.org/10.1038/d41586-020-01282-z.
Marek, Scott, Brenden Tervo-Clemmens, Finnegan J. Calabro, David F. Montez, Benjamin P. Kay, Alexander S. Hatoum, Meghan Rose Donohue, et al. 2022.
“Reproducible Brain-Wide Association Studies Require Thousands of Individuals.” Nature 603 (7902): 654–60.
https://doi.org/10.1038/s41586-022-04492-9.
Martin, L., K. Bülau, C. Hüttl, R. A. Schmitt, D. Perdikis, L. Domide, H. Taher, L. Stefanovski, and P. Ritter. 2024. “Semantic Annotation of Computational Neuroscience Knowledge: The Virtual Brain Ontology for Reproducible Whole-Brain Models.”
Sanz-Leon, Paula, Stuart A. Knock, Andreas Spiegler, and Viktor K. Jirsa. 2015.
“Mathematical Framework for Large-Scale Brain Network Modeling in the Virtual Brain.” NeuroImage 111 (May): 385–430.
https://doi.org/10.1016/j.neuroimage.2015.01.002.
Schild, H. H. 1992.
MRI Made Easy (... Well Almost). Berlex Laboratories.
https://books.google.de/books?id=mG5rAAAAMAAJ.
Schirner, Michael, Gustavo Deco, and Petra Ritter. 2023.
“Learning How Network Structure Shapes Decision-Making for Bio-Inspired Computing.” Nature Communications 14 (1).
https://doi.org/10.1038/s41467-023-38626-y.
Schirner, Michael, Anthony Randal McIntosh, Viktor Jirsa, Gustavo Deco, and Petra Ritter. 2018.
“Inferring Multi-Scale Neural Mechanisms with Brain Network Modelling.” eLife 7 (January): e28927.
https://doi.org/10.7554/elife.28927.
Schirner, Michael, Simon Rothmeier, Viktor K. Jirsa, Anthony Randal McIntosh, and Petra Ritter. 2015.
“An Automated Pipeline for Constructing Personalized Virtual Brains from Multimodal Neuroimaging Data.” NeuroImage 117 (August): 343–57.
https://doi.org/10.1016/j.neuroimage.2015.03.055.
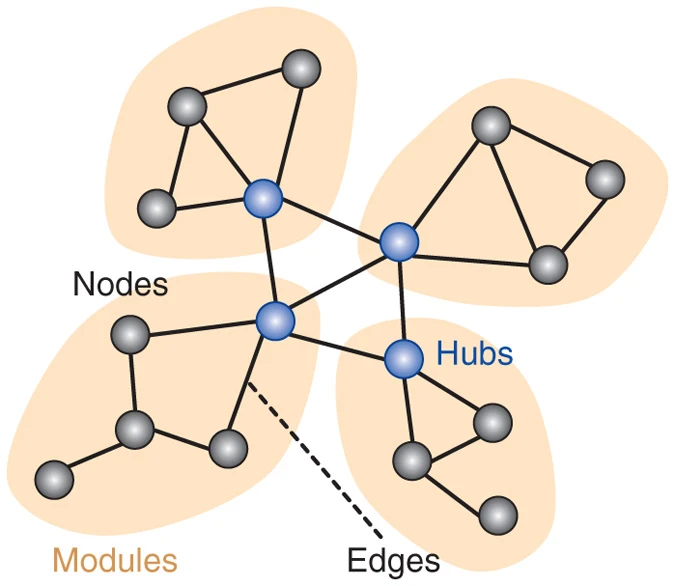
Sporns, Olaf. 2013.
“Making Sense of Brain Network Data.” Nature Methods 10 (6): 491–93.
https://doi.org/10.1038/nmeth.2485.
———. 2014.
“Contributions and Challenges for Network Models in Cognitive Neuroscience.” Nature Neuroscience 17 (5): 652–60.
https://doi.org/10.1038/nn.3690.
Wang, Huifang E, Marmaduke Woodman, Paul Triebkorn, Jean-Didier Lemarechal, Jayant Jha, Borana Dollomaja, Anirudh Nihalani Vattikonda, et al. 2022.
“Virtual Epileptic Patient (VEP): Data-Driven Probabilistic Personalized Brain Modeling in Drug-Resistant Epilepsy.” medRxiv, January, 2022–01.
https://doi.org/10.1101/2022.01.19.22269404.
Zorzos, Ioannis, Ioannis Kakkos, Errikos M. Ventouras, and George K. Matsopoulos. 2021.
“Advances in Electrical Source Imaging: A Review of the Current Approaches, Applications and Challenges.” Signals 2 (3): 378–91.
https://doi.org/10.3390/signals2030024.